Resources & Support
Protocols - Microarray
Microarray technology enables the simultaneous detection and quantification of a large number of proteins or nucleic acids in a variety of biological samples, including cells, tissues, and bodily fluids. The term microarray refers to a miniaturized solid support or chip, such as a nylon membrane or glass microscope slide, which is spotted with a capture element (antibody, protein, oligonucleotide, cDNA, etc.) in a grid-like configuration. There are many applications of microarrays including assessing protein families, probing serum for diagnostic information, profiling expression in response to drugs, and profiling cells treated with certain stimulators or transfected with a given gene. Although there are many procedural variations, the five essential steps in the preparation of a microarray are:
1) capture element synthesis,2) preparation of the solid support surface,
3) immobilization of the capture elements onto the solid support (using a robotic arrayer),
4) binding of the target molecule to the immobilized capture elements, and
5) detection and quantification of the target/capture element complex.
Antibody microarrays use antibodies, raised against known protein antigens, as the capture elements and proteins capable of specifically binding to the immobilized antibodies as the detected target molecules. Fluorescent dyes such as Cy3, Cy5, FITC, and Texas Red, or enzyme systems such as biotin-horseradish peroxidase, are a sensitive means of detection and visualization. Antibody microarrays exploit many of the same principles applied to well established immunoassay procedures such as ELISA or Western blot; however, microarrays offer many advantages including an increased number of data points (which has a direct effect on accuracy), the ability to detect many proteins simultaneously, and reduced antibody and sample volume requirements. In addition, once an array has been designed and the components validated, a robotic arrayer can rapidly spot limitless copies for use with large sample groups.
In this article, two antibody microarray formats will be highlighted: direct labeling and fluorescence-linked immunosorbent assay (FLISA). Both assays use specific antibodies as the capture element. In direct labeling, all proteins in the sample mixture are labeled prior to capture in contrast to FLISA, where target proteins from an untreated sample mixture are captured and subsequently detected through the use of a biotinylated antibody and a fluorophore-labeled secondary antibody.
Antibodies used in microarray assays work best when they are highly purified, exhibit minimal cross-reactivity, and have been shown to work well in traditional ELISA formats. Initially, antibody solutions are prepared at a predetermined optimal concentration (usually 300-500µg/ml), and then they are assembled into a print plate or microtiter plate, which is used by a robotic arrayer to prepare a spotted solid support. Six to ten replicates per antibody are usually sufficient and, depending on the researcher's needs, up to 500 different antibodies per array can be spotted and immobilized for comparison. The spotted support is blocked by treatment with a BSA solution for at least one hour, and then, the support is ready to be exposed to the samples of interest (serum, urine, cell lysate, etc.)
Figure I depicts direct labeling detection. All proteins in a complex mixture are conjugated to either a flurophore (Figure I-A) or biotin prior to incubation with the capture element. The arrays are then washed and scanned for data analysis. The comparison of signal intensities between samples can be used to measure the relative amounts or expression levels of detected proteins. Alternatively, the comparison of signal intensities to calibrated protein standards, labeled with distinct fluorphores, allows exact expression levels to be quantitatively measured (Figure I-B).
Advantages of this method are its simplicity and the requirement of only one specific antibody per target. The primary disadvantage is increased background signal resulting from the labeling of all proteins in the sample mixture (e.g. albumin in serum). Detection sensitivity is limited by the concentration of the background protein relative to the target protein and is typically around 100ng/ml in serum.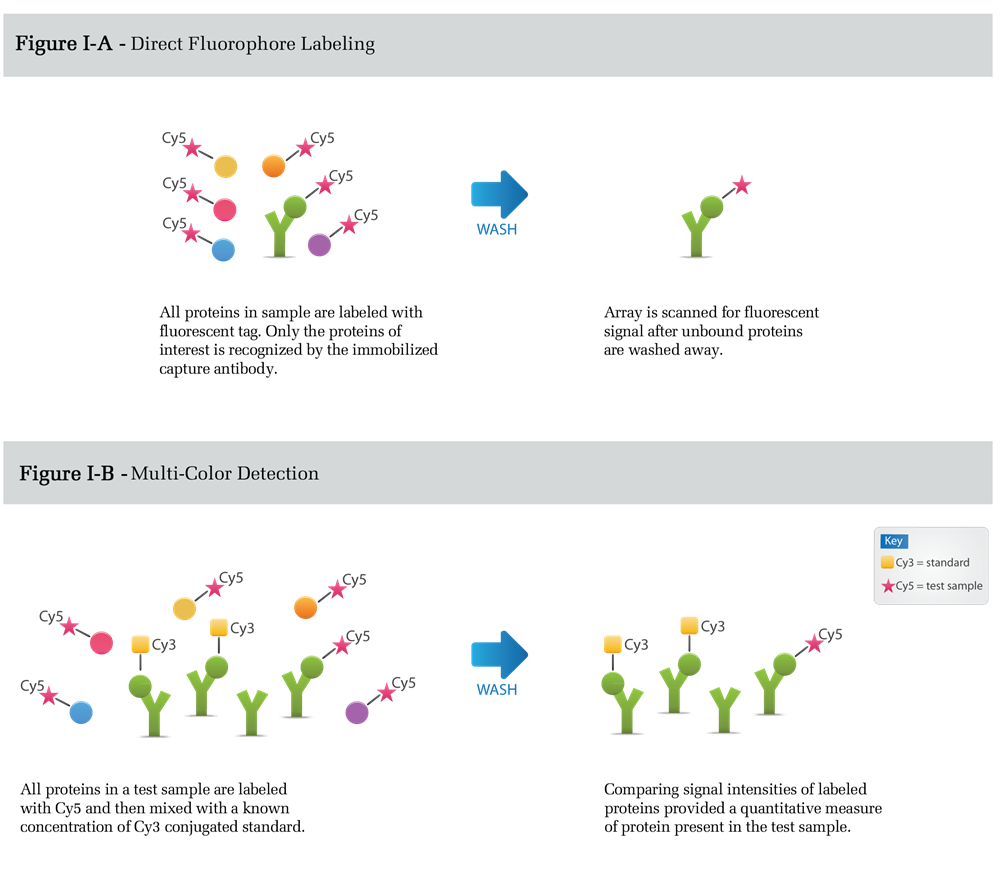
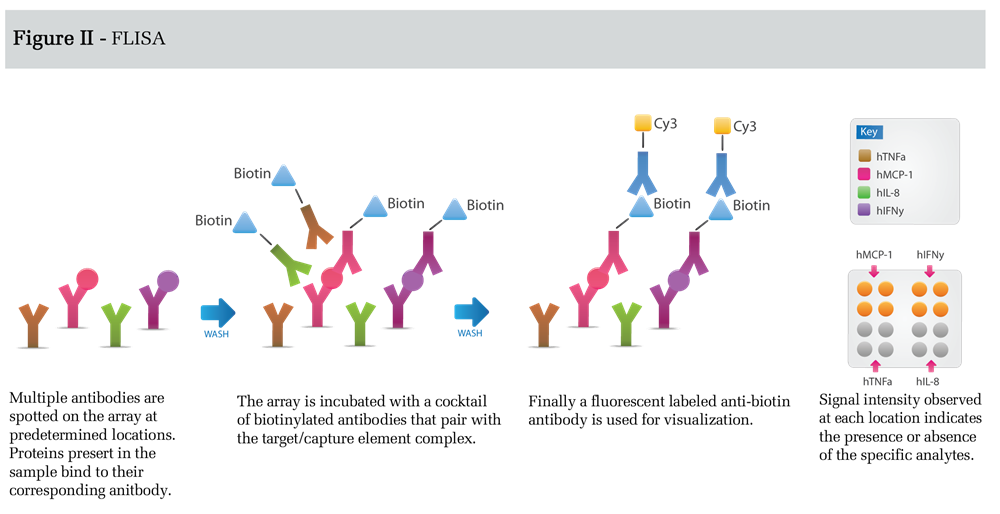
In FLISA detection (Figure II), antibodies spotted on the microarray capture unlabeled protein targets.
This is followed by incubation with a cocktail of biotin-labeled antibodies, each antibody corresponding to one of the spotted capture antibodies. Finally, the array is incubated with a suitable detection antibody such as a fluorophore labeled anti-biotin secondary antibody. The FLISA detection format results in increased sensitivity, relative to the direct labeling method, because background is significantly reduced. Sensitivity in this format can be enhanced even further using tyramide signal amplification or rolling-circle amplification (3).
However, optimization of assays measuring many targets can be difficult using this format; therefore, FLISA detection is usually used for a limited number of targets that are present at concentrations below the detection limit of the direct labeling method. Although the detection limits for the assay depends on a number of parameters, including the affinity and specificity of the antibodies used, the protein background in the sample, and the detection conditions chosen, the FLISA method can provide detection limits in the low pg/ml range.